WRF4L
There is a far more powerful tool to manage work-flow of WRF4G.
Lluís developed a less powerful one which is here described
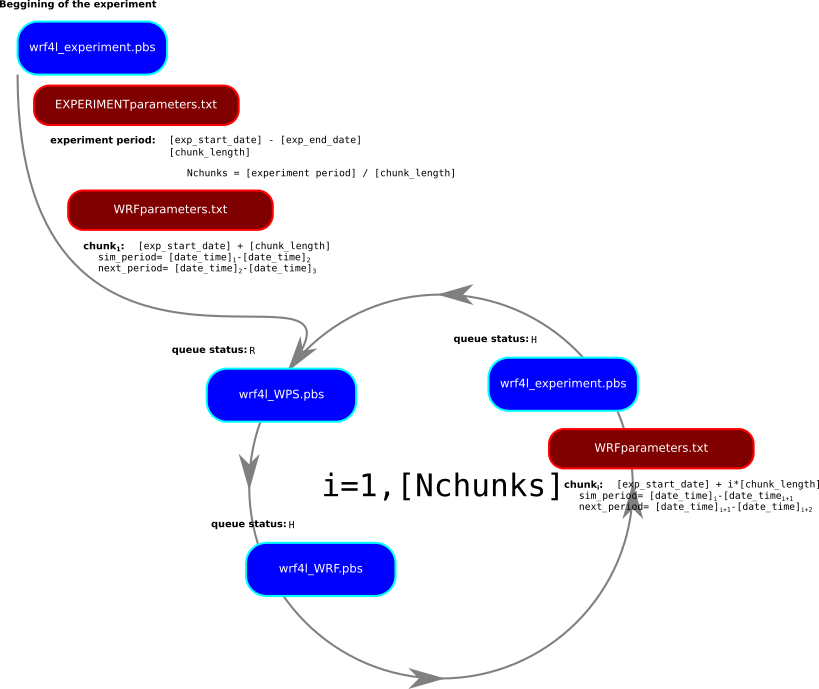
WRF work-flow management is done via 5 scripts (these are the specifics for hydra [CIMA cluster]):
EXPERIMENTparameters.txt: General ASCII file which configures the experiment and chain of simulations (chunks). This is the unique file to modifywrf4l_experiment.pbs: PBS-queue job which prepares the experiment of the environmentwrf4l_WPS.pbs: PBS-queue job which launch the WPS section of the model:ungrib.exe,metgrid.exe,real.exewrf4l_WRF.pbs: PBS-queue job which launch thewrf.exe- There is a folder called
componentswith shell and python scripts necessary for the work-flow management
An experiment which contains a period of simulation is divided by chunks small pieces of times which are manageable by the model. The work-flow follows these steps using run_experiments.pbs:
- Copy and link all the required files for a given chunk of the whole period of simulation following the content of
EXPERIMENTparameters.txt - Launches
wrf4l_WPS.pbswhich will produce the necessary files for the period of the given chunk - Launches
wrf4l_WRF.pbswhich will simulated the period of the given chunk (which waits until the end ofrun_WPS.pbs) - Launches the next
wrf4l_experiments.pbs(which waits until the end ofwrf4l_WRF.pbs)
All the scripts are located in hydra at:
/share/tools/workflows/WRF4L/hydra
How to simulate
- Assuming that we have already defined the domains of simulation and that we have a suited
namelist.wpsandnamelist.inputWRF namelist files in a folder called$GEOGRID. We will gather all this information in the storage folder for the experiment (let's assume called $STORAGEdir) and we create there a folder for the information of the domains.
mkdir ${STORAGEdir}/domains
cp ${GEOGRID}/geo_em.d* ${STORAGEdir}/domains
cp ${GEOGRID}/namelist.wps ${STORAGEdir}/domains
cp ${GEOGRID}/namelist.input ${STORAGEdir}/domains
- We then move to the
${STORAGEdir}/domainsand create a template file for thenamelist.wps
cd ${STORAGEdir}/domains
cp namelist.wps namelist.wps_template
diff namelist.wps namelist.wps_template
4,6c4,6
< start_date = '2010-06-01_00:00:00', '2010-06-01_00:00:00'
< end_date = '2010-06-16_00:00:00', '2010-06-16_00:00:00'
< interval_seconds = 10800,
---
> start_date = 'iyr-imo-ida_iho:imi:ise', 'iyr-imo-ida_iho:imi:ise',
> end_date = 'eyr-emo-eda_eho:emi:ese', 'eyr-emo-eda_eho:emi:ese',
> interval_seconds = infreq
- In case we are going to use
nc2wpsto generate the intermediate files (ungrib.exeoutput directly generated from netCDF files), we need to generate a template for thenamelist.nc2wps. There are different options available in/share/tools/workflows/WRF4L/hydra. In this example we use the one prepared for ERA5 data as it is kept in Papa-Deimos:
cp /share/tools/workflows/WRF4L/hydra/namelist.nc2wps_template_ERA5 ${STORAGEdir}/domains/namelist.nc2wps_template
- Creation of a new folder from where launch the experiment [ExperimentName] (g.e. somewhere at
salidas/folder)
$ mkdir salidas/[ExperimentName]
cd salidas/[ExperimentName]
- copy basic WRF4L files to this folder
cp /share/tools/workflows/WRF4L/hydra/EXPERIMENTparameters.txt ./
cp /share/tools/workflows/WRF4L/hydra/launch_experiment.pbs ./
- Edit the configuration/set-up of the simulation of the experiment (e.g. period of simulation, version of WPS and WRF to use, additional
namelist.input parameters, multiple folders: storageHOME, runHOME, MPI configuration, ...)
$ vim EXPERIMENTparameters.txt
- Change the name of the user in the
launch_experiment.pbs
diff launch_experiment.pbs /share/tools/workflows/WRF4L/hydra/launch_experiment.pbs
11c11
< #PBS -M lluis.fita@cima.fcen.uba.ar
---
> #PBS -M [user]@cima.fcen.uba.ar
- Launch the experiment
$ qsub launch_experiment.pbs
When it is running one would have (runnig WRF job wrf_[SimName] `R', and exp_[SimName] in hold `H'):
$ qstat -u $USER
hydra:
Req'd Req'd Elap
Job ID Username Queue Jobname SessID NDS TSK Memory Time S Time
-------------------- -------- -------- ---------------- ------ ----- --- ------ ----- - -----
397.hydra lluis.fi larga wps_ 27567 1 16 20gb 168:0 R --
398.hydra lluis.fi larga wrf_ -- 1 1 20gb 168:0 H --
399.hydra lluis.fi larga exp_ -- 1 1 2gb 168:0 H --
In case of crash of the simulation, after fixing the issue, go to [runHOME]/[ExpName]/[SimName] and re-launch the experiment (after the first run the scratch is switched automatically to `false')
$ qsub wrf4l_experiment.pbs
Checking the experiment
Once the experiment runs, one needs to look on (following name of the variables from EXPERIMENTparameters.txt
[runHOME]/[ExpName]/[SimName]: Will content the copies of the templates namelist.wps, namelist.input and a file chunk_attemps.inf which counts how many times a chunk has been attempted to be run (if it reached 4 times, the WRF4L is stopped)[runHOME]/[ExpName]/[SimName]/run: actual folder where the computing nodes run the model. In a folder called wrfout there is a folder for each chunk with the standard output of the model[runHOME]/[ExpName]/[SimName]/run/outwrf/[YYYYi][MMi][DDi][HHi][MIi][SSi]-[YYYYf][MMf][DDf][HHf][MIf][SSf]: folder with the standard output and all the required files to run a given chunk. The content of all this folder is compressed and kept in [storageHOME]/[ExpName]/[SimName]/config_[YYYYi][MMi][DDi][HHi][MIi][SSi]-[YYYYf][MMf][DDf][HHf][MIf][SSf].tar.gz[storageHOME]/[ExpName]/[SimName] (in [storageHOST]): output of the already ran chunks as [YYYYi][MMi][DDi][HHi][MIi][SSi]-[YYYYf][MMf][DDf][HHf][MIf][SSf] for a chunk from [YYYYi]/[MMi]/[DDi] [HHi]:[MIi]:[SSi] to [YYYYf]/[MMf]/[DDf] [HHf]:[MIf]:[SSf]
When something went wrong
If there has been any problem check the last chunk (in outwrf/[PERIODchunk]) to try to understand what happens and where the problem comes from:
rsl.[error/out].[nnnn]: These are the files which content the standard output while running the model. One file for each process. If the problem was something related to model execution and it has been prepared for the error, a correct message must appear. (look first for the largest files...
$ ls -lrS rsl.error.*
run_wrf.log: These are the files which content the standard output of the model. Search for `segmentation faults' in form of (it might differ):
forrtl: error (63): output conversion error, unit -5, file Internal Formatted Write
Image PC Routine Line Source
wrf.exe 00000000032B736A Unknown Unknown Unknown
wrf.exe 00000000032B5EE5 Unknown Unknown Unknown
wrf.exe 0000000003265966 Unknown Unknown Unknown
wrf.exe 0000000003226EB5 Unknown Unknown Unknown
wrf.exe 0000000003226671 Unknown Unknown Unknown
wrf.exe 000000000324BC3C Unknown Unknown Unknown
wrf.exe 0000000003249C94 Unknown Unknown Unknown
wrf.exe 00000000004184DC Unknown Unknown Unknown
libc.so.6 000000319021ECDD Unknown Unknown Unknown
wrf.exe 00000000004183D9 Unknown Unknown Unknown
- on
[runHOME]/[ExpName]/[SimName], check the output of the PBS jobs. Which are called:
exp-[SimName].o[nnnn]: output of the wrf4l_experiment.pbswps-[SimName].o[nnnn]: output of the wrf4l_WPS.pbswrf-[SimName].o[nnnn]: output of the wrf4l_WRF.pbs
- Check
[runHOME]/[ExpName]/[SimName]/run/namelist.output which holds all the parameters (even the default ones) used in the simulation
EXPERIMENTSparameters.txt
This ASCII file configures all the simulation. It assumes:
- Required files, forcings, storage, compiled version of the code might be at different machines.
- There is a folder with a given template version of the
namelist.input which will be used and changed accordingly to the requirement of the experiments
Location of the WRF4L main folder (example for hydra)
# Home of the WRF4L
wrf4lHOME=/share/workflows/WRF4L
Name of the machine where the experiment is running (NOTE: a folder with specific work-flow must exist as $wrf4lHOME/${HPC})
# Machine specific work-flow files for $HPC (a folder with specific work-flow must exist as $wrf4lHOME/${HPC})
HPC=hydra
Name of the compiler used to compile the model (NOTE: a file called $wrf4lHOME/arch/${HPC}_${compiler}.env must exist)
# Compilation (a file called $wrf4lHOME/arch/${HPC}_${compiler}.env must exist)
compiler=intel
Name of the experiment
# Experiment name
ExpName = WRFsensSFC
Name of the simulation. Here is understood that a given experiment could have the model configured with different set-ups (here identified with a different name of simulation)
# Simulation name
SimName = control
Which binary of python 2.x to be used
# python binary
pyBIN=/home/lluis.fita/bin/anaconda2/bin/python2.7
Should this simulation be run from the beginning or not. If it is set to `true', it will remove all the pre-existing content of the folder [ExpName]/[SimName] in the running and in the storage spaces. Be careful. In case of `false' simulation will continue from the last successful ran chunk (checking the restart files).
# Start from the beginning (keeping folder structure)
scratch = false
Period of the simulation of the simulation (In this example from 1958 Jan 1st to 2015 Dec 31)
# Experiment starting date
exp_start_date = 19790101000000
# Experiment ending date
exp_end_date = 20150101000000
Length of the chunks (do not make chunks larger than 1-month!!)
# Chunk Length [N]@[unit]
# [unit]=[year, month, week, day, hour, minute, second]
chunk_length = 1@month
Selection of the machines and users to each machine where the different requirement files are located and the output should be placed.
- NOTE: this will only work if one set-up the
.ssh public/private keys in each involved USER/HOST.
- NOTE 2: All the forcings, compiled code, ... are already at
hydra at the common space called share
- NOTE 3: From the computing nodes, one can not access to the
/share folder and to any of the CIMA's storage machines: skogul, freyja, ... For that reason, one need to use these system of [USER]@[HOST] accounts. *.pbs scripts uses a series of wrappers of the standard functions: cp, ln, ls, mv, .... which manage them `from' and `to' different pairs of [USER]@[HOST]. NOTE: This will only work if the public/private ssh key pairs have been set-up (see more details at llaves_ssh)
# Hosts
# list of different hosts and specific user
# [USER]@[HOST]
# NOTE: this will only work if public keys have been set-up
##
# Host with compiled code, namelist templates
codeHOST=lluis.fita@hydra
# forcing Host with forcings (atmospherics and morphologicals)
forcingHOST=lluis.fita@hydra
# output Host with storage of output (including restarts)
outHOST=lluis.fita@hydra
Templates of the configuration of WRF: namelist.wps, namelist.input files. NOTE: they will be changed according to the content of EXPERIMENTparameters.txt like period of the chunk, atmospheric forcing, differences of the set-up, ... (located in the [codeHOST]
# Folder with the `namelist.wps', `namelist.input' and `geo_em.d[nn].nc' of the experiment
domainHOME = /home/lluis.fita/salidas/estudios/dominmios/SA50k
Folder where the WRF model will run in the computing nodes (on top of that there will be two more folders [ExpName]/[SimName]). WRF will run at the folder [ExpName]/[SimName]/run
# Running folder
runHOME = /home/lluis.fita/estudios/WRFsensSFC/sims
Folder with the compiled version of the WPS (located at [codeHOST])
# Folder with the compiled source of WPS
wpsHOME = /share/WRF/WRFV3.9.1/ifort/dmpar/WPS
Folder with the compiled version of the WRF (located at [codeHOST])
# Folder with the compiled source of WRF
wrfHOME = /share/WRF/WRFV3.9.1/ifort/dmpar/WRFV3
Folder to storage all the output of the model (history files, restarts and compressed file with content of the configuration and the standard output of the given run). The content of the folder will be organized by chunks (located at [storageHOST])
# Storage folder of the output
storageHOME = /home/lluis.fita/salidas/estudios/WRFsensSFC/sims/output
Wether modules should be load (not used for hydra)
# Modules to load ('None' for any)
modulesLOAD = None
Names of the files used to check that the chunk has properly ran
# Model reference output names (to be used as checking file names)
nameLISTfile = namelist.input # namelist
nameRSTfile = wrfrst_d01_ # restart file
nameOUTfile = wfrout_d01_ # output file
Extensions of the files with the configuration of WRF (to be retrieved from codeHOST and domainHOME)
# Extensions of the files with the configuration of the model
configEXTS = wps:input
To continue from a previous chunk one needs to use the `restart' files. But they need to be renamed, because otherwise they will be re-written. Here one specifies the original name of the file [origFile] and the name to be used to avoid the re-writting [destFile]. It uses a complex bash script which even can deal with the change of dates according to the period of the chunk (':' list of [origFile]@[destFile]). They will located at the [storageHOST]
# restart file names
# ':' list of [tmplrstfilen|[NNNNN1]?[val1]#[...[NNNNNn]?[valn]]@[tmpllinkname]|[NNNNN1]?[val1]#[...[NNNNNn]?[valn]]
# [tmplrstfilen]: template name of the restart file (if necessary with [NNNNN] variables to be substituted)
# [NNNNN]: section of the file name to be automatically substituted
# `[YYYY]': year in 4 digits
# `[YY]': year in 2 digits
# `[MM]': month in 2 digits
# `[DD]': day in 2 digits
# `[HH]': hour in 2 digits
# `[SS]': second in 2 digits
# `[JJJ]': julian day in 3 digits
# [val]: value to use (which is systematically defined in `run_OR.pbs')
# `%Y%': year in 4 digits
# `%y%': year in 2 digits
# `%m%': month in 2 digits
# `%d%': day in 2 digits
# `%h%': hour in 2 digits
# `%s%': second in 2 digits
# `%j%': julian day in 3 digits
# [tmpllinkname]: template name of the link of the restart file (if necessary with [NNNNN] variables to be substituted)
rstFILES=wrfrst_d01_[YYYY]-[MM]-[DD]_[HH]:[MI]:[SS]|YYYY?%Y#MM?%m#DD?%d#HH?%H#MI?%M#SS?%S@wrfrst_d01_[YYYY]-[MM]-[DD]_[HH]:[MI]:[SS]|YYYY?%Y#MM?%m#DD?%d#HH?%H#MI?%M#SS?%S
Folder with the input data (located at [forcingHOST]).
# Folder with the input morphological forcing data
indataHOME = /share/DATA/re-analysis/ERA-Interim
Format of the input data and name of files
# Data format (grib, nc)
indataFMT= grib
# For `grib' format
# Head and tail of indata files names.
# Assuming ${indataFheader}*[YYYY][MM]*${indataFtail}.[grib/nc]
indataFheader=ERAI_
indataFtail=
In case of netCDF input data, there is a bash script which transforms the data to grib, to be used later by ungrib
Variable table to use in ungrib
# Type of Vtable for ungrib as Vtable.[VtableType]
VtableType=ERA-interim.pl
Folder with the atmospheric forcing data (located at [forcingHOST]).
# For `nc' format
# Folder which contents the atmospheric data to generate the initial state
iniatmosHOME = ./
# Type of atmospheric data to generate the initial state
# `ECMWFstd': ECMWF 'standard' way ERAI_[pl/sfc][YYYY][MM]_[var1]-[var2].grib
# `ERAI-IPSL': ECMWF ERA-INTERIM stored in the common IPSL way (.../4xdaily/[AN\_PL/AN\_SF])
iniatmosTYPE = 'ECMWFstd'
Here on can change values on the template namelist.input. It will change the values of the provided parameters with a new value. If the given parameter is not in the template of the namelist.input it will be automatically added.
## Namelist changes
nlparameters = ra_sw_physics;4,ra_lw_physics;4,time_step;180
Name of WRF's executable (to be localized at [orHOME] folder from [codeHOST])
# Name of the exectuable
nameEXEC=wrf.exe
':' separated list of netCDF file names from WRF's output which do not need to be kept
# netCDF Files which will not be kept anywhere
NokeptfileNAMES=''
':' separated list of headers of netCDF file names from WRF's output which need to be kept
# Headers of netCDF files need to be kept
HkeptfileNAMES=wrfout_d:wrfxtrm_d:wrfpress_d:wrfcdx_d
':' separated list of headers of restarts netCDF file names from WRF's output which need to be kept
# Headers of netCDF restart files need to be kept
HrstfileNAMES=wrfrst_d
Parallel configuration of the run.
# WRF parallel run configuration
## Number of nodes
Nnodes = 1
## Number of mpi procs
Nmpiprocs = 16
## Number of shared memory threads ('None' for no openMP threads)
Nopenthreads = None
## Memory size of shared memory threads
SIZEopenthreads = 200M
## Memory for PBS jobs
MEMjobs = 30gb
Generic definitions
## Generic
errormsg=ERROR -- error -- ERROR -- error
warnmsg=WARNING -- warning -- WARNING -- warning